International Journal of Cancer and Clinical Research
Prdx Overexpression in Tumor Tissue of Breast Cancer Patients
Jillian Muhlbauer1 and Shelley A Phelan2*
1Harvard School of Dental Medicine, Longwood Avenue Boston, USA
2Department of Biology, Fairfield University, Fairfield, USA
*Corresponding author:
Shelley A. Phelan, PhD, Department of Biology, Fairfield University, 1073 North Benson Road, Fairfield, CT 06824, USA, Tel: 203-254-4000, Extn: 3120, E-mail: sphelan@fairfield.edu
Int J Cancer Clin Res, IJCCR-3-053, (Volume 3, Issue 2), Original Research Article; ISSN: 2378-3419
Received: February 04, 2016 | Accepted: April 06, 2016 | Published: April 08, 2016
Citation: Muhlbauer J, Phelan SA (2016) Prdx Overexpression in Tumor Tissue of Breast Cancer Patients. Int J Cancer Clin Res 3:053. 10.23937/2378-3419/3/2/1053
Copyright: © 2016 Muhlbauer J, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Peroxiredoxin (Prdx) proteins are evolutionarily conserved thiol-specific antioxidant enzymes that reduce various cellular peroxides, protecting cells from oxidative damage. Prdxs also have been demonstrated to play an important role in regulating redox-sensitive cell signaling in a number of cell processes. Prdxs have been implicated in cancer biology, and are upregulated in many cancers including breast cancer, as well as several breast cancer cell lines. To explore the entire Prdx family in breast cancer, we analyzed the expression of all six Prdx proteins in breast tumor tissue and adjacent normal breast tissue from 14 patients. We found that most patients have a marked elevation of Prdx expression in tumor tissue, with many patients overexpressing multiple Prdxs. Specifically, we found that 71% of patients overexpress Prdx1, 50% overexpress Prdx2, 64% overexpress Prdx3, and 57% overexpress Prdx4. In contrast, Prdx5 and Prdx6 are elevated in a minority of patients. We found no association, however, between the incidence of Prdx overexpression and tumor grade in this study. Our findings provide further evidence for Prdx elevation in breast cancer using matched patient samples, and support the notion that Prdx upregulation may provide a survival advantage to breast cancer cells, and other cancer cells.
Keywords
Peroxiredoxins, Prdx, Breast Cancer, Tumor, Antioxidant
Introduction
The Peroxiredoxins are a family of evolutionarily conserved thiol-specific antioxidant proteins. These proteins act as peroxidase enzymes, reducing cellular peroxides (including hydroperoxides and lipid peroxides) in the presence of specific electron donors [1]. As a result, peroxiredoxins (Prdxs) protect cells from oxidative damage associated with normal cellular metabolism [2,3]. Since ROS are now well characterized signaling molecules, Prdxs are also recognized to act as modulators of redox signaling, thereby regulating a number of redox-sensitive processes involved in cell proliferation, differentiation, and apoptosis [4,5]. In mammals, there are six Prdx proteins, which can be subcategorized by the number of cysteines in the active site of the protein. The 2-Cys Prdxs (Prdx1-5) react with peroxides to form a disulfide bond either as dimers (Prdx1-4) or intramolecularly (Prdx5) [6]. In contrast, 1 -Cys Prdx (Prdx6) contains only one active cysteine, cannot form dimers through disulfide bond formation, and has the unique ability to reduce lipid peroxides [7]. The different Prdxs exhibit differences in tissue distribution, cellular localization, and substrate specificity, therefore demonstrating distinct roles in the cellular antioxidant defense system [8].
In recent years, the overexpression of Peroxiredoxins in breast cancer cells has been well documented [9-13]. Although the precise mechanism of action in these cells has not been fully elucidated, elevation of other antioxidants in cancer cells suggests that upregulation of peroxiredoxins provides a survival advantage to cancer cells in an environment of elevated oxidative stress. Additionally, research implicating oxidative stress in chemoresistance supports peroxiredoxins and other antioxidant proteins as potential candidates for therapy targets [13]. Recent studies have reported Prdx overexpression correlates with development, recurrence, and/or progression of cancers, [13-15]. Karihtala and others reported elevated Prdx 3-5 levels to be associated with more poorly differentiated tumors, and Prdx 5 with shorter survival in breast cancer patients [10]. They hypothesized that the increase in Prdx expression is induced by higher ROS levels in the cancerous state. However, the correlation with malignancy and disease progression remains a controversy as many groups have conflicting results [10,16,17]. Prdx2 and Prdx4 may be of particular interest since they are secreted proteins and may be useful biomarkers measured in serum samples [16]. Researchers are continuing to search for reliable serum biomarkers in breast cancer in the hopes of earlier detection, and thus better disease prognosis [16,17].
Previous research from our lab has shown overexpression of five of the six Prdxs at the mRNA and protein levels in the widely studied MCF-7 human breast cancer cell line, compared to the non-cancerous MCF-10A cell line [18,19]. We further showed that suppression of these proteins in MCF-7 cells results in reduced cell viability [18]. While investigation of the mechanism of Prdx regulation and function continues in these and other cell lines, it is essential to pursue Prdx studies in human tumor samples to better understand its physiological relevance and aberrant in vivo regulation. In addition, the patient-to-patient variability in Prdx expression, and significant evidence pointing to Prdx misregulation as an epigenetic cancer adaption, rather than a causal factor in carcinogenesis, suggests that proper controls for these experiments will be critical to determine when and how these Prdxs modify their regulation. As such, we sought to analyze Prdx protein expression in a number of patient samples by comparing Prdx levels in breast tumor tissue and adjacent normal tissue within each patient.
Materials and Methods
Two OncoPair INSTA-Blots membranes were purchased from Imgenex/Novus (San Diego, CA). These blots were cat # NBP2-29911 (IMB-130a) and NBP2-30127 (IMB-130d). Each ready-to-use membrane contained denatured protein lysates from ductal carcinomas and adjacent tissues from seven patient donors. All patients were females ranging in age from 36-85 years old and differed in the location and stage of breast cancer. As described (Imagenex/Novus), all protein lysates were diluted to 1 mg/ml and equal amounts (14 ug) were loaded onto each lane of a denaturing gel, electrophoresed and transferred to a PVDF membrane along with a molecular weight marker. Gel loading corresponds to the documented information.
Membranes were pre-wet in methanol and rinsed with TBST. They were blocked in 5% milk in TBST for one hour, incubated in primary antibody overnight at 4°C, rinsed 5 × 15 min in TBST, and incubated in the appropriate alkaline phosphatase-conjugated secondary antibody for one hour at room temperature. After additional rinses with TBST, the chemiluminescent CDP-Star Reagent (GE) was used as a substrate, and the blots were imaged with X-OMAT film (Kodak). The primary antibodies used for the analysis were from Abcam: anti-Prdx1 (ab59538), anti-Prdx2 (ab15572), anti-Prdx3 (ab16751), anti-Prdx4 (ab59542), anti-Prdx5 (ab16944) anti-Prdx6 (ab16947) (Abcam, Cambridge, MA).
Band intensities on western blots were quantified with Image J software. Prdx levels were normalized to control levels and relative fold differences between tumor and adjacent normal tissue were calculated for each patient. Correlation coefficients were calculated to compare tumor grade with incidence of Prdx elevation, for each Prdx.
Results
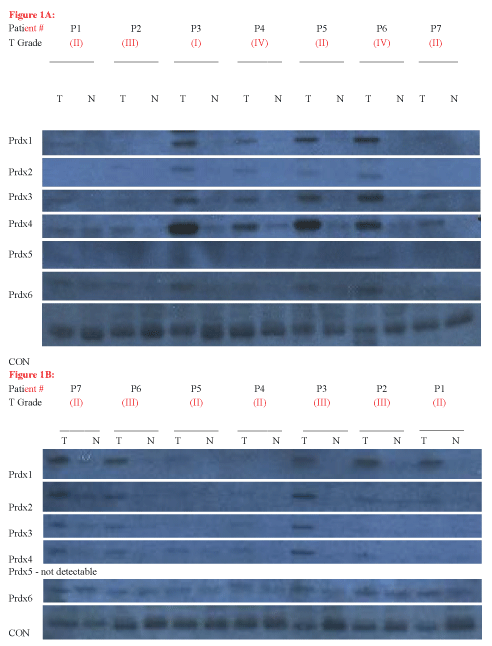
We analyzed two patient blots, obtained from Imgenex/Novus. Each blot contained paired samples of tumor/non-tumor breast tissue from 7 patients, allowing us to analyze a total of 14 patients. Figure 1(A & B) shows the results of this experiment. As shown in Figure 1, almost all of the Prdx proteins were detectable in most of the patients, but at varying levels. Prdx5 was detected at the lowest levels in virtually all patients, and was undetectable on Blot 130a (Figure 1B).
.
Figure 1: Prdx Expression in Tumor (T) and Adjacent Normal (N) Tissue. (A) Oncopair INSTAblot IMB-130d, including matched tumor and adjacent non-tumor breast tissue for 7 patient donors. Tumor diagnosis for patients are as follows: P1, mucinous carcinoma; P2, mucinous carcinoma; P3, mucinous carcinoma; P4, metaplastic; P5, intraductal carcinoma; P6, intraductal carcinoma; P7, intraductal carcinoma; (B) Oncopair INSTAblot IMB-130a, including matched tumor and adjacent non -tumor breast tissue for 7 patient donors. All breast tumors were diagnosed as ductal carcinoma. (*note - the patient orientation of this blot is reversed from the published order, based on a difference in loading order for this blot lot).
View Figure 1
Our experiment revealed tumor overexpression of multiple Prdx proteins in nearly all patients, with one patient exhibiting tumor overexpression of all six Prdx proteins (Figure 1A, P6). In particular, four patients from blot 130d (Figure 1A) and three patients from blot 130a (Figure 1B) exhibited a very substantial increase in the expression of several Prdx proteins in the tumor, compared to adjacent tissue. Interestingly, Prdxs1-4 were overexpressed to the greatest extent in tumors (Figure 1) and also were overexpressed in the greatest number of patient tumors. The tumor-specific elevation was over five-fold in many patients, reaching 15X for Prdx1, 7X for Prdx2, 14X for Prdx3, and 16X for Prdx4. Interestingly, patient #3 (Figure 1A) exhibited the highest elevation of Prdx1, Prdx2 and Prdx4. Prdx6 levels reached 7X, while quantification of Prdx5 levels could not be made due to low signal.
We then quantified the number and percentage of patients who clearly (and unambiguously) showed elevated expression of each Prdx. As shown in Table 1, the first four Prdxs were elevated in 71%, 50%, 64%, and 57% of patients, respectively. Finally, we quantified the number of patients exhibiting overexpression of multiple Prdx proteins. As shown in table 2, 64% of patients overexpressed two or more Prdxs in tumor tissue and 50% of patients overexpressed 4 or more Prdxs in tumor tissue. Of the 14 patients analyzed, we found only three patients who exhibited little to no Prdx elevation in tumor tissue (Figure 1A, P2; Figure1B, P3 & P4).
![]()
Table 1: Percent of Patients having elevated tumor Prdx Levels.
View Table 1
![]()
Table 2: Percent of Patients having elevated tumor Levels of Multiple Prdxs.
View Table 2
Based on prior conflicting studies regarding the relationship between Prdx levels and tumor grade, we wanted to examine this in our samples. The tumor grade for each patient was available from the blot supplier (Imgenex/Novus). We examined whether the incidence of Prdx overexpression correlated with tumor grade in these patient samples. As shown in Figure1, we found Prdx overexpression in all four tumor grades (I, II, III and IV), and we saw no particular relationship between the tumor grade and the incidence of Prdx elevation for any of the Prdx proteins, as determined by correlation analysis.
Discussion
In the present study, we examined the expression of the six Peroxiredoxin antioxidant proteins in tumor tissue and adjacent normal tissue from 14 breast cancer patients. We found that the vast majority of patients show a marked overexpression of multiple Prdx proteins in tumor tissue, with Prdx1, 3 and 4 being most frequently elevated, and by the largest extent. We found no correlation between incidence of Prdx overexpression and tumor grade.
Our findings are consistent with other studies that have examined multiple Prdxs, including studies that specifically found elevated expression of Prdx1-3 in breast tumors [9], and another showing elevation in Prdx1, 3, and 4 in a much higher percentage of breast cancer than either Prdx2 or Prdx6 [10]. However, these other studies used non-matched normal breast tissue as controls. Since Prdx levels vary significantly between individuals, our study demonstrating elevated tumor levels compared to normal breast tissue within the same individual is more compelling, and more strongly supports a physiologically important role for Prdx in the cancer process. Our observations that Prdx1-4 are elevated in most tumors are also consistent with our previous studies showing overexpression of Prdxs1 -5 (but not Prdx6) in the MCF7 breast cancer cell line, as compared to the normal MCF10A line [18,19]. This helps to validate the use of the MCF7/MCF10A cell lines as an acceptable in vitro model for further investigation of Prdx regulation and function in breast cancer. Despite considerable evidence that Prdxs are upregulated in breast cancer, as well as other cancers, there is virtually nothing known about the mechanism of this upregulation in these cells.
Our observation that Prdx1-4 are most commonly elevated in breast tumors is intriguing. A recent review explores new evidence on the mechanism of action of these 2-Cys peroxiredoxins (Prdx1-4), revealing their important role in redox-sensitive processes including proliferation and cancer [20]. Each of these proteins uses a similar dimeric structure to catalyze the peroxide reaction, and has a somewhat distinct subcellular localization. Prdx1 has been the most studied, and is the most abundant Prdx in cells, localizing to the cytosol. Cha et al. reported overexpression of Prdx1 and Thioredoxin I in breast carcinoma, using paired samples, and further reported an association with tumor grade [21]. However, this study compared fold difference in mRNA levels (not protein) between tumor grade and type, and examined a much larger set of samples, so it is possible that we may have observed a correlation with a larger set of samples.
Prdx1 has been found to associate with multiple proteins including c-Abl [22], and recently, Prdx1 was shown to associate with MAPK phosphatases (MKP-1 and MKP-5), thereby regulating redox-signaling in breast cancer in a H202 dose-dependent manner [23]. Based on its dual-role, it is possible that upregulation of Prdx1 in breast cancer either serves to protect cancer cells from ROS-induced damage and/or assists in redox-regulated proliferation and apoptosis. Prdx2 is another cytoplasmic peroxiredoxin, while Prdx3 is the primary mitochondrial peroxiredoxin, which is critical for proper mitochondrial function [24]. Prdx4 is both secreted and found in the ER, and recently has been implicated in the oxidative protein folding [25,26]. While upregulation of all four of these peroxiredoxins has been reported in breast cancer cells, there is little known about their role in these tumor cells. It is clear that each Prdx has designated peroxide-detoxifying roles in the cell, and that levels of ROS in all of these subcellular locations are elevated in cancer cells. A recent report from our lab demonstrated that suppression of Prdxs 1,2,3 or 5 in MCF7 breast cancer cells significantly reduces resistance to doxorubicin, while suppression of Prdx3 reduces cell viability even in the absence of drug treatment [18]. This is the only study, thus far, that has examined the effect of all six Prdx proteins in breast cancer cells.
Much additional research is needed to elucidate the precise role of each Prdx in breast cancer biology, and the mechanism of its upregulation in tumor cells. But together, our findings further implicate overexpression of the Prdx family of proteins in breast cancer as a widespread phenomenon, and support the notion that Prdx induction may provide cytoprotection for breast cancer cells in vivo.
Acknowledgements
This work was supported by research funding provided to the PI by Fairfield University.
References
-
Chae HZ, Chung SJ, Rhee SG (1994) Thioredoxin-dependent peroxide reductase from yeast. J Biol Chem 269: 27670-27678.
-
Rhee SG, Kang SW, Chang TS, Jeong W, Kim K (2001) Peroxiredoxin, a novel family of peroxidases. IUBMB life 52: 35-41.
-
Fujii J, Ikeda Y (2002) Advances in our understanding of peroxiredoxin, a multifunctional, mammalian redox protein. Redox Report 7: 123-130.
-
Immenschuh S, Baumgart-Vogt E (2005) Peroxiredoxins, oxidative stress, and cell proliferation. Antioxid Redox Signal 7: 768-777.
-
Rhee SG, Woo HA, Kil IS, Bae SH (2012) Peroxiredoxin functions as a peroxidase and a regulator and sensor of local peroxides. J Biol Chem 287: 4403-4410.
-
Hall A, Karplus PA, Poole LB (2009) Typical 2‐cys peroxiredoxins–structures, mechanisms and functions. FEBS J 276: 2469-2477.
-
Fisher AB (2011) Peroxiredoxin 6: A bifunctional enzyme with glutathione peroxidase and phospholipase A2 activities. Antioxid Redox Signal 15: 831-844.
-
Wood ZA, Schroder E, Robin Harris J, Poole LB (2003) Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci 28: 32-40.
-
Noh DY, Ahn SJ, Lee RA, Kim SW, Park IA, et al. (2001) Overexpression of peroxiredoxin in human breast cancer. Anticancer Res 21: 2085-2090.
-
Karihtala P, Mantyniemi A, Kang SW, Kinnula VL, Soini Y (2003) Peroxiredoxins in breast carcinoma. Clin Cancer Res 9: 3418-3424.
-
Chahed K, Kabbage M, Hamrita B, Guillier CL, Trimeche M, et al. (2008) Detection of protein alterations in male breast cancer using two dimensional gel electrophoresis and mass spectrometry: The involvement of several pathways in tumorigenesis. Clin Chim Acta 388: 106-114.
-
Chang XZ, Li DQ, Hou YF, Wu J, Lu JS, et al. (2007) Identification of the functional role of peroxiredoxin 6 in the progression of breast cancer. Breast Cancer Res 9: R76.
-
Li DQ, Wang L, Fei F, Hou YF, Luo JM, et al. (2006) Identification of breast cancer metastasis‐associated proteins in an isogenic tumor metastasis model using two‐dimensional gel electrophoresis and liquid chromatography‐ion trap‐mass spectrometry. Proteomics 6: 3352-3368.
-
Lehtonen ST, Svensk AM, Soini Y, Paakko P, Hirvikoski P, et al. (2004) Peroxiredoxins, a novel protein family in lung cancer. Int J Cancer 111: 514-521.
-
Quan C, Cha EJ, Lee HL, Han KH, Lee KM, et al. (2006) Enhanced expression of peroxiredoxin I and VI correlates with development, recurrence and progression of human bladder cancer. J Urol 175: 1512-1516.
-
Liu F J, Wang XB, Cao AG (2014) Screening and functional analysis of a differential protein profile of human breast cancer. Oncol Lett 7: 1851-1856.
-
Misek DE, Kim EH (2011) Protein biomarkers for the early detection of breast cancer. Int J Proteomics 2011: 343582.
-
McDonald C, Muhlbauer J, Perlmutter G, Taparra K, Phelan SA (2014) Peroxiredoxin proteins protect MCF-7 breast cancer cells from doxorubicin-induced toxicity. Int J Oncol 45: 219-226.
-
Tehan L, Taparra K, Phelan S (2013) Peroxiredoxin overexpression in MCF-7 breast cancer cells and regulation by cell proliferation and oxidative stress. Cancer Invest 31: 374-384.
-
Jinah Park, Sunmi Lee, Sanghyuk Lee, Sang Won Kang (2014) 2-cys peroxiredoxins: Emerging hubs determining redox dependency of mammalian signaling networks. Int J Cell Biol 2014: 715867.
-
Cha MK, Suh KH, Kim IH (2009) Overexpression of peroxiredoxin I and thioredoxin1 in human breast carcinoma. J Exp Clin Cancer Res 28: 93.
-
Wen ST, Van Etten RA (1997) The PAG gene product, a stress-induced protein with antioxidant properties, is an Abl SH3-binding protein and a physiological inhibitor of c-Abl tyrosine kinase activity. Genes Dev 11: 2456-2467.
-
Turner-Ivey B, Manevich Y, Schulte J, Kistner-Griffin E, Jezierska-Drutel, et al. (2013) Role for Prdx1 as a specific sensor in redox-regulated senescence in breast cancer. Oncogene 32: 5302-5314.
-
Wonsey DR, Zeller KI, Dang CV (2002) The c-Myc target gene PRDX3 is required for mitochondrial homeostasis and neoplastic transformation. Proc Natl Acad Sci U S A 99: 6649-6654.
-
Tavender TJ, Springate JJ, Bulleid NJ (2010) Recycling of peroxiredoxin IV provides a novel pathway for disulphide formation in the endoplasmic reticulum. EMBO J 29: 4185-4197.
-
Zito E, Melo EP, Yang Y, Wahlander A, Neubert TA, et al. (2010) Oxidative protein folding by an endoplasmic reticulum-localized peroxiredoxin. Mol Cell 40: 787-797.





