Journal of Infectious Diseases and Epidemiology
Prevalence of β-Lactamase Positive Ampicillin Resistant H. Influenzae from Children of Bangladesh
Harunur Rashid1,2*, Shereen Shoma1 and Mahbubur Rahman1
1International Center for Diarrheal Disease Research, Bangladesh
2Department of Cell Biology, University of Alabama at Birmingham, USA
*Corresponding author:
Harunur Rashid, International Center for Diarrheal Disease Research, Bangladesh and Department of Cell Biology, University of Alabama, Birmingham, USA, Tel: +1-205-996-5767, Fax: +12059965109, E-mail: hrashid07@gmail.com
J Infect Dis Epidemiol, JIDE-2-010, (Volume 2, Issue 1), Original Article; ISSN: 2474-3658
Received: November 14, 2015 | Accepted: March 24, 2016 | Published: March 26, 2016
Citation: Rashid H, Shoma S, Rahman M (2016) Prevalence of β-Lactamase Positive Ampicillin Resistant H. Influenzae from Children of Bangladesh. J Infect Dis Epidemiol 2:010. 10.23937/2474-3658/1510010
Copyright: © 2016 Rashid H, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
A total of 255 NPS specimens were collected from every seventh of 1786 children with meningitis (n = 293) and pneumonia (n = 1493). One hundred and twenty one (47.5%) H. influenzae were isolated from 255 NPS specimens. Among 121 isolates, 15 (38%) were recovered from meningitis compared to 106 (49%) from pneumonia cases (P < 0.3). Fifteen percent of isolates (18/121) were serotype b (Hib) and the rest were non-Hib. The rate of resistance to ampicillin, amoxicillin, chloramphenicol, azithromycin and co-trimoxazole revealed by E-test were; 17.3%, 17.3%, 22%, 1% and 55% respectively. All strains were susceptible to ciprofloxacin, moxifloxacin, gatifloxacin, levofloxacin and ceftriaxone. A significant difference in antimicrobial resistance between Hib and non-Hib was noted for ampicillin (67% vs 8.7%; P < 0.0000001), chloramphenicol (72.2% vs 12.6%; P < 0.000001), and co-trimoxazole (88.9% vs 48.5%; P < 0.004). Seventeen percent (21/121) of H. influenzae were ampicillin resistant due to β-lactamase. All ampicillin resistant isolates were characterized as multi-drugs resistant (MDR). Of the 21 ampicillin-resistant H. influenzae, 57% (12/ 21) were Hib, followed by non-capsulated (28.6%), serotype d (9.5%) and serotype c (4.8%). MDR was significantly different (P < 0.0000001) between Hib (66.7%) and non-Hib (8.7%). Biotype II (38.8%), III (20.7%), and I (18.1%) comprise 78% of H. influenzae isolates. Ninety-five percent of Hib belong to biotype I. Thus, 67% of the Hib isolates represent β-lactamase mediated ampicillin and multi-drugs resistance which is a tremendous public health concern.
Keywords
Haemophilus influenzae, β-lactamase, Antimicrobial resistance, Biotyping
Introduction
Haemophilus influenzae is responsible for a number of human diseases ranging from chronic respiratory infection to meningitis. H. influenzae asymptomatically colonizes the nasopharynx of healthy individuals. Colonized H. influenzae isolates are potential to causes systemic disease and mucous membrane infections. Almost all of the children colonized by respiratory pathogens at some time of their life, and about 15% of colonized children infected within a month of colonization [1]. Bacterial species colonized in healthy carrier is easily spread to new hosts and possesses threat of infection in the new host. Therefore, nasopharyngeal carriage plays important role in spread of the potential pathogens. The H. influenzae infection begins with the colonization of the upper respiratory tract which make nasopharyngeal colonization is a critical risk factor for potential infection and progression to disease [2]. Study showed the children frequently colonized by H. influenzae are more likely to develop acute otitis media compared to the children who colonized less frequently [3]. Eight different biotypes and six serotypes are used as epidemiological markers for studying the pattern of colonization with H. influenzae and to identify the pathogenic strain [4]. However, biotype I and serotype b are commonly associated with meningitis in children, and biotypes II and III are commonly associated with upper respiratory tract infections [5,6]. Non-capsulated and biotype IV H. influenzae has been shown as an important pathogen in neonatal, maternal, and female genital infections.
In contrast, non-serotypeable strains of H influenzae, particularly biotypes II and III, are frequently commensal to the upper respiratory tract. While colonization with biotypes II and III usually does not progress to disease. However, sometimes these biotypes have been emerged as pathogen of sinusitis, otitis media, acute and chronic exacerbations of lower respiratory tract infection, and acute and chronic conjunctivitis [7,8]. Continuous monitoring of the antimicrobial resistance patterns of H. influenzae is recommended for cost-effective treatment of invasive infections. Isolation of H. influenzae from cerebrospinal fluid and blood from rural hospital in developing countries may not be possible due to the lack of adequate laboratory facilities. Hence, the monitoring of nasopharyngeal colonization with H. influenzae as a surrogate marker for invasive and locally invasive H. influenzae could be an attractive option [9]. This study was undertaken to investigate serotypes, biotypes and antimicrobial susceptibility patterns of H. influenzae colonized in the nasopharynx of young children as well as study the role of β-lactamase in the development of ampicillin resistance.
Materials and Methods
Study population
A prospective study was conducted on nasopharyngeal colonization by H. influenzae for 36 months from July 2000 to June 2003 in three hospitals in Dhaka, Bangladesh. A total of 255 nasopharyngeal samples (NPS) from every seventh of 1786 pediatric case having pneumonia and meningitis aged less than 5 years were included in this study. Clinical and demographical data were recorded in a questionnaire. The population consisted of children of both sex, aged less than five years. Written consent was obtained from the parents or guardians of all children. The study protocol was approved by the institutional ethical committee of the Centre (ICDDR,B).
Specimen collection
Nasopharyngeal specimen was collected immediately after examination. A calcium alginate tipped swab was used for the collection of nasopharyngeal specimens. The swab was inoculated on chocolate agar plate with bacitracin (300 μgml-1) and immediately transported to the laboratory via cold box and immediately incubated at 37°C in candle extinction jar for 20 to 24 hours.
Microbiological procedure
After overnight incubation the plates were examined for H. influenzae. All respiratory pathogens were isolated and identified by standard laboratory methodologies: colony morphology, Gram staining characteristics, and X (hemin) and V (nicotinamide adenine dinucleotide) factor requirements test. Each isolate was then stocked in trypticase soy broth with 20% glycerol and stored at -80°C for further investigation.
Antimicrobial susceptibility
Antibiotic susceptibility was qualitatively determined by disk diffusion method according to Standard Clinical laboratory Institute (Previously National Committee for Clinical Laboratory Standard; NCCLS) guideline [10]. Minimum inhibitory concentration (MIC) was also determined E-test [11] technique using antimicrobial strips of ampicillin, amoxicillin, azithromycin, trimethoprim-sulfamethoxazole (TMP-SMX), ciprofloxacin, gatifloxacin, moxifloxacin, chloramphenicol, ceftriaxone, (AB Biodisk, Solna, Sweden). Antimicrobial resistance cut-off value was determined using European Committee on Antimicrobial Susceptibility Testing guideline [12]. Haemophilus testing media (HTM) supplemented with X and V factor was used for susceptibility testing of H. influenzae. The plates were incubated in candle extinction jar (~5% CO2) at 37°C for 24 h prior to reading of the results. MICs were interpreted by using Standard Clinical laboratory Institute breakpoints. The MIC was defined as the lowest concentration of antibiotic that inhibited the visible growth. Isolates were considered multi-resistant if they had reduced susceptibility to at least three structurally unrelated antibiotics. H. influenzae ATCC 49247 strains was used for the quality control.
Detection of β-lactamase production
All the ampicillin resistant strains and some ampicillin susceptible strains were tested for β-lactamase production by nitrocefin stick test (Cefinase stick Oxoid, UK).
Serotyping
The strains of H. influenzae were serotyped by the slide agglutination test, using antiserum type a to f (Denka Saiken).
Biotype analysis
Biotypes were investigated to all the H. influenzae isolates by determining their ability to produce urease, indole and ornithine decarboxylase [6,13] using locally prepared media supplemented with NAD (5 μgml-1) and haemin (5 μgml-1).
Statistical analysis
Differences were statistically analyzed using the Chi-square method with the Yates correction as appropriate. P-value of < 0.05 was considered significant.
Results
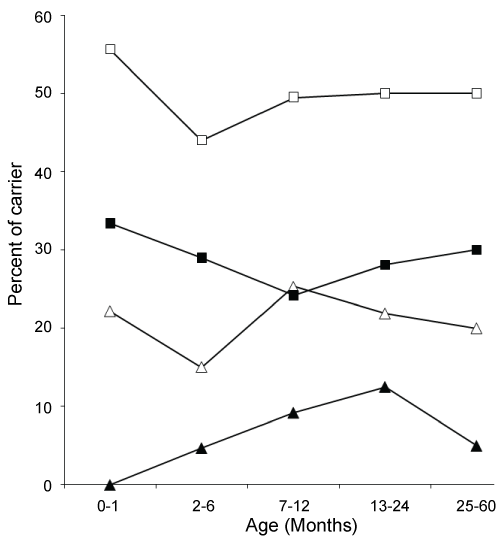
A total of 255 nasopharyngeal samples (NPS) collected from every seventh of 1786 [meningitis (293) and pneumonia (1493)] patients under five year of age. Among 255 NPS specimens, 39 (15%) were from meningitis and 216 (85%) from pneumonia case. Sixty two percent (159/255) of the subjects included in this study were male. A total of 121 (47.5%) strains of H. influenzae were isolated from 255 specimens. Among them 15 (38%) isolates were from meningitis compare to 106 (49%) isolates from pneumonia cases (P < 0.3). The rate of colonization with H. influenzae in male (50%) was higher compare to female (44%) (P < 0.5). About 80% subjects less than 12 months of age and there is no remarkable variation of H. influenzae colonization among different age groups (Figure 1). The rates of resistance to ampicillin, amoxicillin, chloramphenicol, azithromycin and co-trimoxazole were; 17.3%, 17.3%, 18%, 1% and 54% respectively (Table 1). In addition, 3% and 1% strains were intermediately resistant to chloramphenicol and co-trimoxazole respectively. All strains were susceptible to ciprofloxacin, moxifloxacin, gatifloxacin, levofloxacin and ceftriaxone (Table 1). MIC50/MIC90 of Ampicillin, amoxicillin, azithromycin, chloramphenicol, cotrimoxazole, ciprofloxacin, gatifloxacin, levofloxacin, moxifloxacin and ceftriaxone were shown at Table 1. Of the 121 isolates, 18 (14.9%) were H. influenzae type b (Hib). A significant difference in antimicrobial resistance between Hib and non-Hib was observed for ampicillin (67% vs 8.7%; P < 0.0000001), chloramphenicol (72.2% vs 12.6%; P < 0.000001), and co-trimoxazole (88.9% vs 48.5%; P < 0.004) (Table 1).
.
Figure 1: Percent of H. influenzae (HI) total carrier (□), non-typeable HI carrier (■), typeable HI carrier (Δ) and Hib carrier (▲) status of children among different age groups.
View Figure 1
![]()
Table 1: Susceptibility and MIC result of colonized isolates of H. influenza.
View Table 1
All of the ampicillin resistant H. influenzae isolates, 21 (100%) were positive for β-lactamase. Seventeen percent of (21/121) isolates were multi-drugs resistant (MDR; resistant ≥ 3 drugs), and MDR is significantly higher among the Hib (66.7%) and biotype I (50%). Chloramphenicol and co-trimoxazole resistance is significantly higher in ampicillin resistant isolates (81% and 100%) compared to the susceptible strains (9% and 45%) (P < 0.0000001 and P < 0.000005 respectively). The distribution of all biotypes, serotype and ampicillin resistant, is shown in (Table 2). All eight biotypes were found in our H. influenzae bacterial population. Biotype II was most common (38.8 %), followed by III (20.7%) and I (18.1%); cumulatively comprises 78% of H. influenzae isolates. A strong correlation was seen between biotypes and H. influenzae type b (Table 2). H. influenzae, type b is mostly (94.4%) belongs to biotype I, while the non-Hib H. influenzae is more common in the rest of the biotypes especially in II (41%) and III (19%). Most (75%) of the uncapsulated H. influenzae isolates were biotype II (49%) and III (26%). Forty five percent serotype c and 35% serotype d isolates were belonged to biotype II. Of the 21 ampicillin-resistant H. influenzae, 57% (12/21) were Hib followed by non-serotypeable (28.6%), serotype d (9.5%) and serotype c (4.8%). All of the ampicillin resistant H. influenzae isolates belong to the biotypes I (52.4%), II (19%), III (19%) and IV (9.5%). Fifty percent isolates of biotype I was β-lactamase positive ampicillin resistant (BLPAR) followed by 28.5% of biotype IV, 16% of biotype III and 8.5% biotype II. So, BLPAR isolates of H. influenzae are mostly associated with seorotype b (66.7%) and biotype I (50%) which is the major cause of invasive infection in Bangladeshi children.
![]()
Table 2: Biotypes and serotypes of H. influenzae colonizing in the nasopharynx of the children (n = 121).
View Table 2
Discussion
To our knowledge, this is the first Bangladeshi study on prevalence, serotyping, biotyping and antimicrobial susceptibility among H. influenzae colonization in the nasopharynx of children with pneumonia and meningitis. Our study demonstrated a higher rate of nasopharyngeal carriage of H. influenzae (47.5%), especially serotype b (14.9%) and biotype I-III (77.7%). The overall frequency of ampicillin resistant H. influenzae was 17.3%. All the ampicillin resistant strains were β-lactamase positive. Many study conducted in different parts of the world described the carriage rate of H. influenzae at ranging from 11.6 to 76% [9,13-15]. The carriage rate of H. influenzae in our study (47.5%) is higher than the study from India (41.7%; [13]), and England (30.5%; [16]), and lower than the study from Central Asia (57%; [17]).
The carriage rates of H. influenzae and Hib among diseased children were 40.8% vs 0% in Portugal [18], and 32.6% vs 2% in Pakistan [19]. The highest carrier rate of H. influenzae and Hib were 88.3% and 16.4% respectively reported in Australian aboriginal children with acute lower respiratory infections (ALRI) [20]. The carriage rate of Hib in our study (14.9%) is comparable to that in a study from India (13.2%; [13]), and higher than the study from Gambia (12%; [21]), central Asia (11%; [17]), Australia (11%, [20]), Turkey (7.2%; [22]), Thailand (7%; [23]) and England (1.1%; [16]). The rate of non-capsulated H. influenzae among colonized isolates in our study was 57.9% compare to 97.9% in Portugal [18], 79.9% in Australia [20], 91.3% in Pakistan [19], and 92.4% in England [16]. The higher nasopharyngeal carrier rate of H. influenzae in our study can be explained by the fact that all of our study subjects had respiratory illness compared to other studies conducted on healthy children.
The overall frequency of ampicillin resistant H. influenzae was 17.3%. All the ampicillin resistant strains were β-lactamase positive that supported by another Bangladeshi study [24] and a study from Brazil [25]. BLPAR varies considerably with geographical region, the PROTEKT study reported the highest prevalence of ampicillin resistant H. influenzae from South Korea (64.7%) and lowest from Italy (1.8%) [26]. Fifteen percent (18/121) of the isolates were serotype b. Sixty seven percent of Hib and 8.7% (9/103) of non-Hib were ampicillin resistant in our study compared to 44% and 13.1% Hib and non-Hib respectively in India [13]. Our findings of ampicillin resistance is comparable to some study from India (15.8%, [27]; 20 %, [28]), lower than the other Indian studies (21.1%, [29]; 46%, [30]; 79 %, [9] and 22.9%, [13]) higher than the study from Pakistan (4.1%; [19]) and Portugal (10%; [18]). The PROTEKT study [26] has reported a carriage rate of 64.7% for ampicillin-resistant H. influenzae in South Korea, 17.1% in Hong Kong, 8.5% in Japan and 36.4% in a study from United States [31].
Chloramphenicol resistant H. influenzae isolates are still rare in most parts of the world; 0.2-1.9% in USA [31,32] and 4.7% in Turkey [33]. Chloramphenicol resistant H. influenzae are reported by different groups; from 0% [27] to 60% (IBIS study 2002) in India and 34.4% in Pakistan [19].
The highest prevalence of co-trimoxazole resistant H. influenzae is reported from India (30-80%) [9,29,30,34] followed by Pakistan (77.5%; [19]). Other parts of the world reported low rates of co-trimoxazole resistant H. influenzae; 20-50% from other Asian countries [33,35,36] and < 20% in USA and European countries [14,31,36]. Our finding of chloramphenicol and co-trimoxazole resistance (21.5% and 55%) was consistent to the study of India and Pakistan.
A strong correlation between serotype b and biotype I was observed. Serotype b is the pathogenic H. influenzae and biotype I causes mostly invasive diseases like meningitis in children, while biotypes II and III are more commonly associated with upper respiratory tract infections like otitis media and conjunctivitis [4,6]. Our observation coincided with the previous observations [4,6]. We found 77.3% isolates of biotype I belonged to serotype b H. influenzae. Das et al. [9] found that the most common biotype was biotype I (36 %), followed by II (18 %) and III (18 %). In contrast we observed that biotype II (38.8%), was most common followed by biotype III (20.7%), and I (18.1%) cumulatively biotype I, II and III comprises 77.7% of H. influenzae in our study similar to a report by Gratten et al. [20] showed 78% of the H. influenzae colonized isolates were belong to biotype I (20.1%), II (28.3%) and III (30.7%). Our findings on biotype is different from the Indian study conducted by Jain et al. [13] reported that 61% isolates of H. influenzae was biotype I (19.6%), II (16.8%) and III (25%). Another Australian study conducted by Harper and Tilse [37] reported that 90.5% isolates of H. influenzae were belonged to biotype I (15.7%), II (48.5%) and III (26.3%). Most (94.4%) (17/18) of H. influenzae type b was included in biotype I in our study was supported by the observation of Gratten et al. [20] reported that 89.3% (25/28) of Hib was biotype I.
A strong correlation between biotype and ampicillin resistance was also found. All ampicillin resistant isolates of H. influenzae of our study were belonged to biotype I (52.5%), II (19%), III (19%) and IV (9.5%) comparable to the study from India [13] and Brazil [38]. Dabernat et al. [39] reported that 50% of encapsulated strains were biotype I, and often resistant to ampicillin (38.5 %), while 42% of non-encapsulated strains were biotype II with 10.6% resistant to ampicillin. On the other hand, 39.2% (20/51) encapsulated strains were biotype I and 55% (11/20) of them were resistant to ampicillin, while 48.6% (34/70) of non-encapsulated strains were biotype II with 11.8% (4/34) resistant to ampicillin. Similar to our findings, Granato et al. [40] reported that encapsulated strains of biotype I had the highest frequency of ampicillin resistance. No strain was found to be ampicillin resistant without producing β-lactamase in contrast 14.4% (33/229) ampicillin resistant H. influenzae isolates were negative for β-lactamase from Indian study [13].
Overall carriage rate of H. influenzae was consistently remained around 50% in various age groups of the children. However, rate of Hib colonization varied among the different age groups (Figure 1). During the first month of age the carriage rate of Hib was 0%, due to the maternal natural immunization with Hib that provide higher levels of antibody until 2 months of age of babies born to immunized mother [41]. So, Hib vaccination during pregnancy could be protective to newborn by decreasing Hib colonization and the risk of infection. Hib carriers in the age group 2-6 months were all aged > 4-6 months and only a single case of Hib colonization was found above 24 months of age. This can be explained as; after 3 months of age maternal antibody to Hib disappeared in infants and Hib begin to colonize and culminate between 4 and 18 months. Children begin to produce antibody to Hib at around 2 years of age and subsequent colonization with Hib decreased significantly. Colonization pattern of non-Hib encapsulated H. influenzae were similar to Hib except during infancy, colonization with non-Hib encapsulated H. influenzae was higher (22.2%) compare to Hib (0%) (Figure 1). Colonization with non-typeable isolates was significantly differed from the encapsulated H. influenzae (Figure 1).
In our study, high rate of ampicillin (17%), chloramphenicol (21%) and co-trimoxazole (55%) resistance among colonized isolates of H. influenzae reflect the resistance rate of these drugs among the invasive isolates resistant to ampicillin (31%), chloramphenicol (42%), trimethoprim-sulfamethoxazole (44%) and azithromycin (1.4%). BLPAR isolates of H. influenzae of our study were mostly associated with Hib (66.7%) and biotype I (50%) are come out to be most pathogenic causing invasive disease such as pneumonia, meninigitis etc. [4,6,42]. Moreover, higher rate of MDR in H. influenzae to commonly used first line drugs made the treatment of invasive disease very difficult. Therefore, physician has to depend on third generation cephalosporin for the empirical therapy but this drug is expensive and is not affordable by the majority of rural Bangladeshi people. As an alternative, fluoroquinolone such as moxifloxacin and gatifloxacin could be used as empirical therapy. Azithromycin also could be a drug of choice for empirical therapy especially for the adult. Physician should be careful to prescribe azithromycin for the children due to the possible side effect. However, immunization with protein conjugate Hib vaccine is the best option to prevent colonization as well as possible invasive Hib infection among the young children.
Acknowledgements
This work was supported by the United States Agency for International Development, Washington, DC, USA. We are grateful to LSD Team of icddrb and physicians for their assistance during the study.
References
-
Faden H, Duffy L, Wasielewski R, Wolf J, Krystofik DA, et al. (1997) Relationship between nasopharyngeal colonization and the development of otitis media in children. J Infect Dis 175: 1440-1445
-
Unicomb L, Bishop R (1989) Epidemiology of rotavirus strains infecting children throughout Australia during 1986-1987: a study of serotype and RNA electropherotype. Arch Virol 106: 23-34.
-
Ryan MJ, Ramsay M, Brown D, Gay NJ, Farrington CP, et al. (1996) Hospital admissions attributable to rotavirus infection in England and Wales. J Infect Dis 174 Suppl 1: S12-18.
-
Alrawi AM, Chern KC, Cevallos V, Lietman T, Whitcher JP, et al. (2002) Biotypes and serotypes of Haemophilus influenzae ocular isolates. Br J Ophthalmol 86: 276-277.
-
Pittman M (1931) Variation and type specificity in the bacterial species hemophyilus influenzae. J Exp Med 53: 471-492.
-
Kilian M (1976) A taxonomic study of the genus Haemophilus, with the proposal of a new species. J Gen Microbiol 93: 9-62.
-
Murphy TF (1992) Haemophilus influenzae and viruses 100 years later: from influenza to human immunodeficiency virus. Am J Med 92: 583-586.
-
Wald ER (1997) Conjunctivitis in infants and children. Pediatr Infect Dis J 16: S17-20.
-
Das BK, Arora NK, Mathur P, Ostwal P, Mandal S, et al. (2002) Nasopharyngeal carriage of Haemophilus influenzae. Indian J Pediatr 69: 775-777.
-
National Committee for Clinical Laboratory Standards (NCCLS) (2000). NCCLS Performance Standard for Antimicrobial Susceptibility Testing, document M 1000-S13. Wayne, PA: NCCLS.
-
Jorgensen JH, Howell AW, Maher LA (1991) Quantitative antimicrobial susceptibility testing of Haemophilus influenzae and Streptococcus pneumoniae by using the E-test. J Clin Microbiol 29: 109-114.
-
(2016) European Committee on Antimicrobial Susceptibility Testing Breakpoint tables for interpretation of MICs and zone diameters Version 6.0.
-
Jain A, Kumar P, Awasthi S (2006) High ampicillin resistance in different biotypes and serotypes of Haemophilus influenzae colonizing the nasopharynx of healthy school-going Indian children. J Med Microbiol 55: 133-137.
-
Talon D, Leroy J, Dupont MJ, Bertrand X, Mermet F, et al. (2000) Antibiotic susceptibility and genotypic characterization of Haemophilus influenzae strains isolated from nasopharyngeal specimens from children in day-care centers in eastern France. Clin Microbiol Infect 6: 519-524.
-
Uraz G, Simsek H, Celik B (2000) Beta-lactamase activities and resistance to antibiotics of Haemophilus influenzae, H. parainfluenzae and H. aphrophilus strains identified in throat cultures from children. Drug Metabol Drug Interact 16: 217-228.
-
Howard AJ, Dunkin KT, Millar GW (1988) Nasopharyngeal carriage and antibiotic resistance of Haemophilus influenzae in healthy children. Epidemiol Infect 100: 193-203.
-
Factor SH, LaClaire L, Bronsdon M, Suleymanova F, Altynbaeva G, et al. (2005) Streptococcus pneumoniae and Haemophilus influenzae type B Carriage, Central Asia. Emerg Infect Dis 11: 1476-1479.
-
Neto AS, Lavado P, Flores P, Dias R, Pessanha MA, et al. (2003) Risk factors for the nasopharyngeal carriage of respiratory pathogens by Portuguese children: phenotype and antimicrobial susceptibility of Haemophilus influenzae and Streptococcus pneumoniae. Microb Drug Resist 9: 99-108.
-
Mastro TD, Nomani NK, Ishaq Z, Ghafoor A, Shaukat NF, et al. (1993) Use of nasopharyngeal isolates of Streptococcus pneumoniae and Haemophilus influenzae from children in Pakistan for surveillance for antimicrobial resistance. Pediatr Infect Dis J 12: 824-830.
-
Gratten M, Manning K, Dixon J, Morey F, Torzillo P, et al. (1994) Upper airway carriage by Haemophilus influenzae and Streptococcus pneumoniae in Australian Aboriginal children hospitalised with acute lower respiratory infection. South Asian J Trop Med Public Health 25: 123-131.
-
Adegbola RA, Secka O, Lahai G, Lloyd-Evans N, Njie A, et al. (2005) Elimination of Haemophilus influenzae type b (Hib) disease from The Gambia after the introduction of routine immunisation with a Hib conjugate vaccine: a prospective study. Lancet 366: 144-150.
-
Poyrazoglu S, Kömeç S, Gökçay G, Ongen B (2005) Haemophilus influenzae type b carriage among 3- to 24-month-old Turkish children. Epidemiol Infect 133: 1113-1117.
-
Olsen SJ, Dejsirilert S, Sangsuk L, Chunsutiwat S, Dowell SF (2005) Frequent Haemophilus influenzae type B colonization in rural Thailand. Pediatr Infect Dis J 24: 739-742.
-
Saha SK, Baqui AH, Darmstadt GL, Ruhulamin M, Hanif M, et al. (2005) Invasive Haemophilus influenzae type B diseases in Bangladesh, with increased resistance to antibiotics. J Pediatr 146: 227-233.
-
de Almeida AE, de Filippis I, Ferreira DG, de Abreu AO, Rebelo C, et al. (2006) Antimicrobial susceptibility of Haemophilus influenzae isolates collected from 4 centers in Brazil (1990-2003). Diagn Microbiol Infect Dis 54: 57-62.
-
Kohno S, Hoban D, The PROTEKT Surveillance Study Group (2003) Comparative in vitro activity of telithromycin and ß-lactam antimicrobials against bacterial pathogens from community-acquired respiratory tract infections: data from the first year of PROTEKT (1999-2000). J Chemother 15: 335-341.
-
Ayyagari A, Sharma P, Chakraborti A, Agarwal KC (1985) Isolation and detection of Haemophilus influenzae from patients of respiratory tract infections and their antibiotic susceptibility patterns in Chandigarh. Indian J Chest Dis Allied Sci 27: 230-235.
-
Patwari AK, Bisht S, Srinivasan A, Deb M, Chattopadhya D (1996) Aetiology of pneumonia in hospitalized children. J Trop Pediatr 42: 15-20.
-
Nag VL, Ayyagari A, Venkatesh V, Ghar M, Yadav V, et al. (2001) Drug resistant Haemophilus influenzae from respiratory tract infection in a tertiary care hospital in north India. Indian J Chest Dis Allied Sci 43: 13-17.
-
Invasive Bacterial Infections Surveillance (IBIS) Group of the International Clinical Epidemiology Network (2002) Are Haemophilus influenzae infections a significant problem in India? A prospective study and review. Clin Infect Dis 34: 949-957.
-
Doern GV, Brueggemann AB, Pierce G, Holley HP Jr, Rauch A (1997) Antibiotic resistance among clinical isolates of Haemophilus influenzae in the United States in 1994 and 1995 and detection of ß-lactamase-positive strains resistant to amoxicillinclavulanate: results of a national multicenter surveillance study. Antimicrob Agents Chemother 41: 292-297.
-
Johnson DM, Sader HS, Fritsche TR, Biedenbach DJ, Jones RN (2003) Susceptibility trends of Haemophilus influenzae and Moraxella catarrhalis against orally administered antimicrobial agents: five-year report from SENTRY antimicrobial surveillance program. Diagn Microbiol Infect Dis 47: 373-376.
-
Gur D, Ozalp M, Sumerkan B, Kaygusuz A, Töreci K, et al. (2002) Prevalence of antimicrobial resistance in Haemophilus influenzae, Streptococcus pneumoniae, Moraxella catarrhalis and Streptococcus pyogens: results of a multicentre study in Turkey. Int J Antimicrob Agents 19: 207-211.
-
Jain A, Kumar P, Awasthi S (2005) High nasopharyngeal carriage of drug resistant Streptococcus pneumoniae and Haemophilus influenzae in North Indian schoolchildren. Trop Med Int Health 10: 234-239.
-
Hsueh PR, Liu YC, Shyr JM, Wu T, Yan J, et al. (2000) Multicenter surveillance of antimicrobial resistance of Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis in Taiwan during the 1998-1999 respiratory seasons. Antimicrob Agents Chemother 44: 1342-1345.
-
Sahm DF, Jones ME, Hickey ML, Diakun DR, Mani SV, et al. (2000) Resistance surveillance of Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis isolated in Asia and Europe, 1997-1998. J Antimicrob Chemother 45: 457-466.
-
Harper JJ, Tilse MH (1991) Biotypes of Haemophilus influenzae that are associated with noninvasive infections. J Clin Microbiol 29: 2539-2542.
-
Casagrande ST, Vicente EJ, Landgraf IM, Kobata AM (2000) Antimicrobial resistance patterns of Haemophilus influenzae isolated from patients with meningitis in São Paulo, Brazil. Braz J Med Biol Res 33: 295-300.
-
Dabernat H, Delmas C, Rich C, Livrelli V, Joly B (1988) Characterization of Haemophilus influenzae isolated from patients with otitis media. Eur J Clin Microbiol Infect Dis 7: 219-225.
-
Granato PA, Jurek EA, Weiner LB (1983) Biotypes of Haemophilus influenzae: relationship to clinical source of isolation, serotype, and antibiotic susceptibility. Am J Clin Pathol 79: 73-77.
-
Englund JA, Glezen WO, Turner C, Harvey J, Thompson C, et al. (1995) Transplacental antibody transfer following maternal immunization with polysaccharide and conjugate Haemophilus influenzae type b vaccines. J Infect Dis 171: 99-105.
-
Rahman M, Hossain S, Baqui AH, Shoma S, Rashid H, et al. (2008) Haemophilus influenzae type-b and non-b-type invasive diseases in urban children (< 5 years) of Bangladesh: implications for therapy and vaccination. J Infect 56: 191-196.





